Revisiting jupyter notebooks
There has been a lot of discussion about jupyter notebooks in the online channels I consume and the point of this post is to bring them together.
Coming from R and beeing a heavy user of Rmarkdown files jupyter notebooks felt familiar right away but also a bit awkward. Datacamp made the effort of comparing the to feature by feature in a blog post at the end of 2016. It is a bit out-dated but skimming through it most of it still holds to be true. With some exceptions. of course since there have been some developments on both sides. But more on that later
In the end what we want from both tools are four things:
- literate programming, meaning we can have natural language code comments and code output right next to our actual code and have all three components well seperated.
- create awesome code-enriched html content
- polyglot coding, the division of code in small executable chunks also allows us to run each chunk in a different programming language
- comofortable programming, being able to use IDE features, like code completion, visual representation of all variables, data and variable exploration, git-based version control
Both jupyter and Rmarkdown can be used for all these things, however there are some criticism on how jupyter performs in these categories. Most fameously summed up by Joel Grus at jupytercon this year in his presentation and Owain Kenway in ths older blogpost.
To sum this criticism up is that jupyter by design stores its output within the notebook file. The codeoutput can be saved at any time and then code chunks can be executed in any given order. When opening a jupyter notebook, there is no garantuee that execution of all code chunks will again result in the same output. Code output is saved as a binary addition to the underlying json a format that is not compatible with version control tools.
Personally for me this is the reason why I am not using jupyter for programming. Unless I want to create some python-based html content as for this blog for example.
However Brian Granger from project jupyter hase recently been on the dataframed podcast and has talked about his vision of project jupyter. He mentioned to things that stuck with me.
- do not use jupyter notebooks use jupyter lab, because jupyter lab is more advanced and makes things better for you on the IDE side.
- at netflix they are running thousands of jupyter jobs a day.
So this made me very curious because it sounds like finally there is a well implemented solution to ensuring execution of code chunks in order. So I stumbled across this blog post in which netflix was showing their jupyter system landscape.
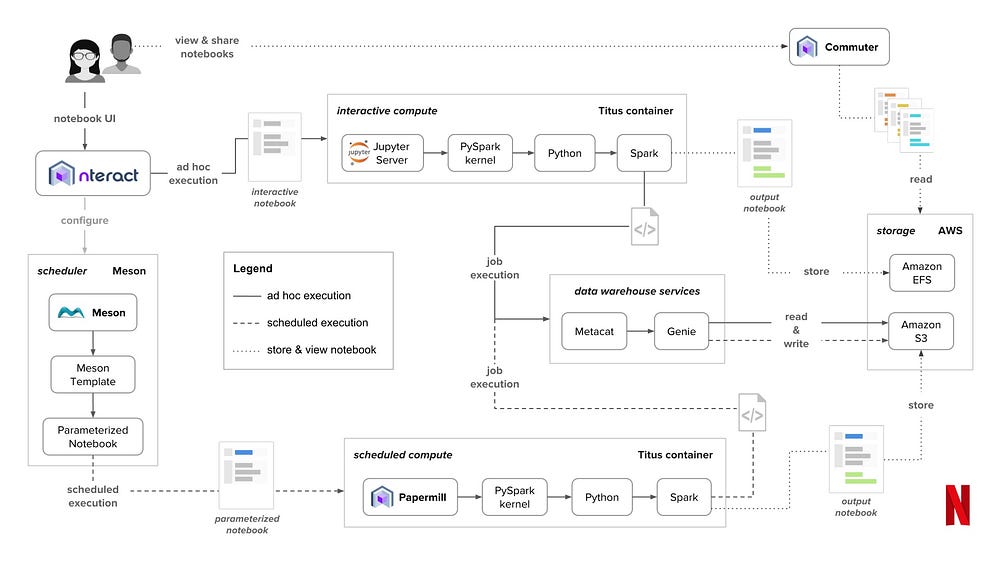
This is quite outstanding, especially the tool papermill caught my attention which allows you to pass parameters to notebooks. A feature I was dearly missing from Rmarkdown. The code output of a jupyter notebook run with a fixed set of parameters via papermill is sure to be reproducible. Also it makes jupyter notebooks production compatible so we do not have to copy paste all the notebook code into a .py file and rewrite the whole thing.
On a second note the company nteract also develops hydrogen one of the most popular atom packages. (atom is an open-source text editor which has become my new favourite IDE for python development)
Parametrized reports are a feature that has been around in Rmarkdown for a while, but it might be difficult to set-up a similar R-Spark landscape that works as well as the python-Spark infrastructure.
So I guess purists will probably always feel a bit cranky about jupyter and its json-based file format. But there seems to be a silver lining and we might finally be able to integrate jupyter more sensibly into our workflows.
But still as Brian Granger mentioned on the dataframed podcast there are no best-practice guidelines for jupyter notebooks at the moment and that is something that they are working on.
